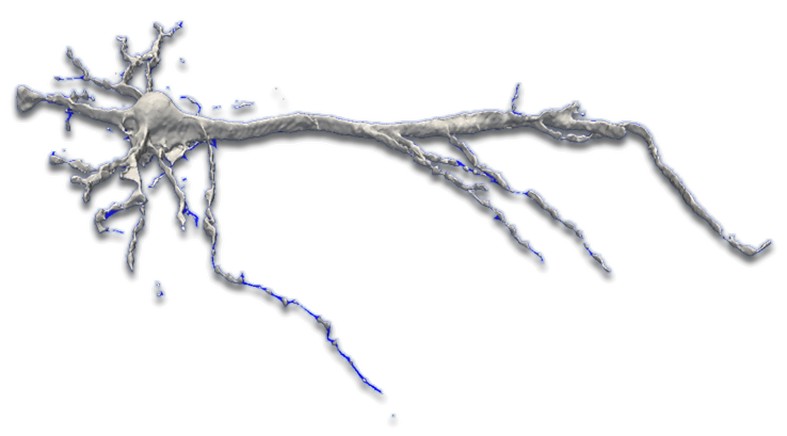
Reconstruction of 3D neuronal structures.
Kanuj KumarAbstract
The three-dimensional reconstruction of neurons is crucial for accurate neurobiological analysis. However, biologists have to rely on time-consuming manual or semi-manual methods to process the laser-scanned microscopy images into geometrical models that in turn would lead to the creation of libraries of neuronal morphologies. Current techniques for digitizing neuronal morphology in 3D entail manual tracing using commercial packages such as NeuroZoom and NeuroLucida. Such methods introduce systematic inaccuracies depending upon the individual performing the tracing, thus, necessitating the need of fully-automated reconstruction methods for neuronal structures. Despite the recent advancements, the automation of reconstruction process either doesn't exhibit robustness against noise of microscopy images or fail to capture precise dendritic structures. Due to these con icting requirements, a perfect reconstruction is impossible making some user intervention necessary. In this thesis, we present a framework for reconstruction of neurons that is substantiated through the user validation of output from each process within framework. The adaptive approaches outlined in this thesis were put to use with the goal of enabling automated and exible outputs to ensure a high quality reconstruction.
Our framework also consists of methods to handle discontinuities in the microscopy image to produce a connected geometrical model and is capable of working on varying contrast and size of data.[PDF]
Supplementary Material:
- Reconstruction of 3D Neuronal Structures - Power Point presentation (Download)