RobustCavities - A software tool for computation for biomolecular cavities in uncertain data.
Cavities refer to empty spaces enclosed by the protein molecule. Existing methods to compute the cavities are sensitive to inaccuracies in the empirically determined atomic positions and radii. We propose a novel notion of robust cavities which are stable under small perturbations in atomic radii, and describe an efficient method of their computation based on alpha shape framework of cavity extraction. A web-server interface RobustCavities is implemented to facilitate interactive computation of robust cavities in proteins. This enables the user to reduce, if not completely eliminate, the inaccuracies in cavity detection due to uncertainty.
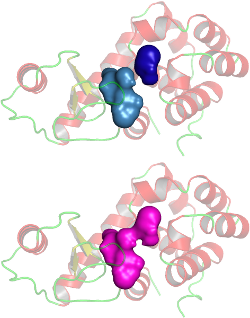
The two cavities that appear very near to each other in a lyzozyme protein (PDB id: 200L) may be a single cavity. The solid surface represents cavities while the protein is shown as cartoon to provide context.
For more details, pease visit the web-server home-page here.
References
- Raghavendra Sridharamurthy, Talha Bin Masood, Harish Doraiswamy, Siddharth Patel, Raghavan Varadarajan and Vijay Natarajan.
Extraction of robust voids and pockets in proteins.
In Visualization in Medicine and Life Sciences III.
Lars Linsen, Hans-Christian Hege, and Bernd Hamann (Eds.)
Springer-Verlag, Mathematics and Visualization Series, 2016, 329-349.
Cover Image - Talha Bin Masood and Vijay Natarajan.
An integrated geometric and topological approach to connecting cavities in biomolecules.
PacificVis 2016: Proc. IEEE Pacific Visualization Symposium, 2016, 104-111.