Topological Analysis Reveals Multiple Pathways in Molecular Dynamics.
Luca Donati, Surahit Chewle, Dominik St Pierre, Vijay Natarajan, and Marcus Weber.Journal of Chemical Theory and Computation, 21(20), 2025, 10385-10397.
[ACS Publications Link]
Abstract
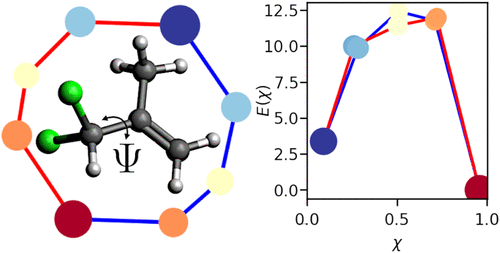
Molecular Dynamics simulations are indispensable tools for comprehending the dynamic behavior of biomolecules, yet extracting meaningful molecular pathways from these simulations remains challenging due to the vast amount of high dimensional data. In this work, we present Molecular Kinetics via Topology (MoKiTo), a novel approach that combines the ISOKANN algorithm to determine the membership function of a molecular system with a topological analysis tool inspired by the Mapper algorithm. Our strategy efficiently identifies and characterizes distinct molecular pathways, enabling the detection and visualization of critical conformational transitions and rare events. This method offers deeper insights into molecular mechanisms, facilitating the design of targeted interventions in drug discovery and protein engineering.[PDF]