Q1 : How is protein input supplied to the ChExVis server?
Ans: ChExVis requires PDB structure of proteins in text format. The PDB structure can be uploaded manually, or just the PDB id can be specified. ChExVis automatically downloads the specified PDB structure from RCSB database. If user has specified computation of trasmembrane channels, then ChExVis gives preference to PDB structure downloaded from OPM database. In cases when OPM does not contain the specified structure, the RCSB structure is used for further computations.
Q2 : Can ChExVis handle biological assemblies of protien structures?
Ans: Yes. However, preferably these should be uploaded manually. If only the PDB id is specified, ChExVis may download the single bio-unit from RSCB database, instead of downloading assembly structure.
Q3 : Is there an upper limit on PDB file size?
Ans: Yes. Files larger than 5MB are not allowed to be uploaded for security reasons.
Q4 : I don't understand all the parameters in job submission form.
Ans: Hovering over the feilds in submission form displays a brief description of that parameter. You can leave them unchanged if you are not sure, because these parameters are already set to good default values.
Q5 : I don't want to give my email-id to this website. Can I still view the results?
Ans: Yes, you can. Specifying email-id is optional. After submission of your job, link to the results page will appear. You can bookmark that link and view the results when they are ready. Specifying your email-id would be helpful in cases when the web-server has a long queue of jobs, resulting in long waiting period.
Q6 : The bio-molecule submitted by me is known to have a channel, but ChExVis does not show me the channel I expected. Why?
Ans: The most common reason for this is that the channel may be very constricted and thus not successfully extracted with default probe radius of 1.00Å. So, if the channel radius goes below 1.00Å at any point along its length, the channel would be considered as infeasible and not reported by ChExVis.
A good solution is to reduce the probe radius to a value less than 1.00Å, you can even reduce it to 0.00Å to ensure channels of any width are considered feasible. If you still don't get expected results, try increasing the number of end-points used while submitting the job, e.g. Auto end points, Inside points and Outside points fields can be set to higher values (say 15 or 20).
In most cases, by doing these adjustments, correct channels would be found and reported as top channels by ChExVis.
Q7 : My results are successfully computed, but I don't see any channels in JMol view and the channel properties screen is blank.
Ans: This is normal behaviour. On top left side of your results page, you will see 3 tables viz. "Channels to active sites", "Pores" and "Transmembrane Pores" (or some of these tables depending on your input). You have to click on some row of these tables to view that particular channel. You can view only one channel at a time.
Q8 : How can I download PyMOL script for the computed channels?
Ans: You can download PyMOL script for each channel individually. You will see a link "Download PyMOL script for this channel" in JMol view window, which will allow you to download the script for the channel currently being viewed. Downloading single PyMOL script for all channels is not supported, this feature would be added in future.
Q9 : Channel Properties Java applet not loading, blocked by JRE, security warnings, etc.
Ans: First make sure that you have latest Java plug-in installed in your browser. We recommend Oracle JRE. After installing the latest JRE, you should restart your browser. Security warnings about Channel Properties applet may still be displayed. We would like to assure you that our code does not contain any malware, the security warning is
displayed by default because the applet is currently self-signed, and not signed by CA. We would resolve this issue in future by obtaining certificate from a trusted CA. In case, your browser completely refuses to run self-signed applets, you have to reduce the Java security level. Go to Java Control panel, and set security level to "Medium". It is set to "High" by default under which self-signed
applets are not allowed to execute. After setting the security to "Medium", the browser will successfully load the applet after displaying a few warning message boxes.
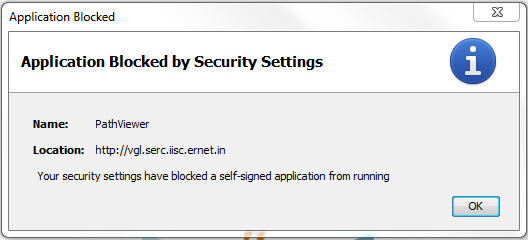
The error message displayed if the applet is blocked by Java security.
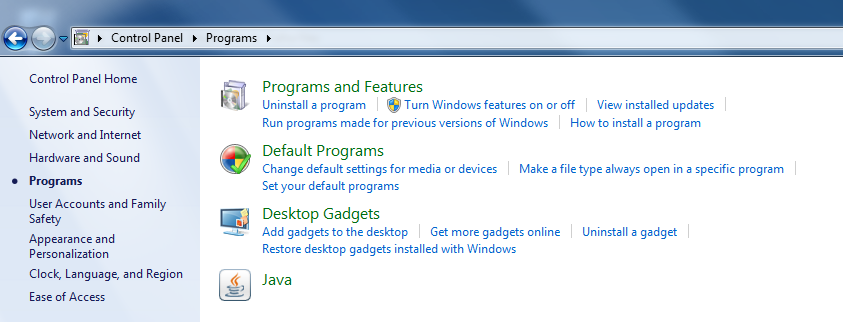
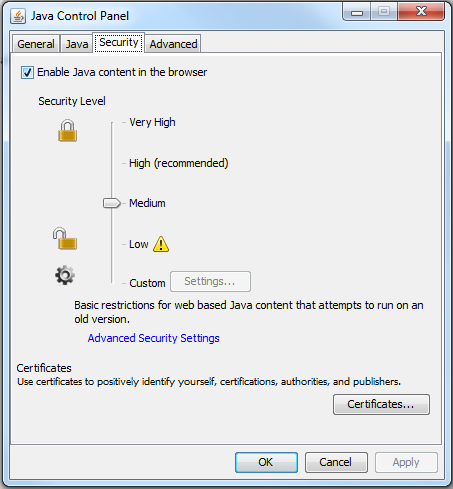
To solve this problem, reduce the Java security level from "High" to "Medium" in Java Control Panel.
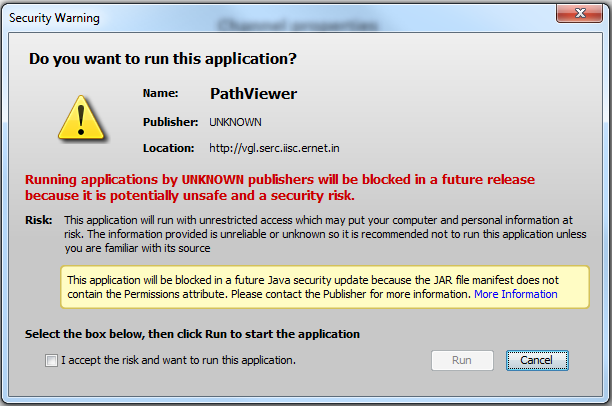
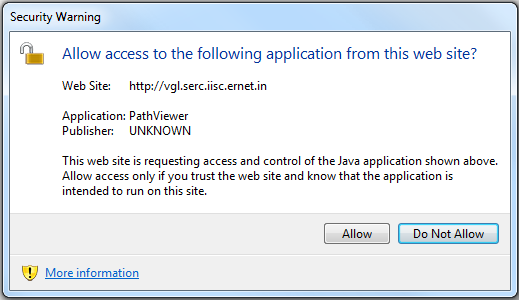
Even at reduced security level, you may see these warning messages. Click "Run" and "Allow" everytime they pop up.