Method Overview
Abstract
Voids and pockets in a protein, collectively called as cavities, refer to empty spaces that are enclosed by the protein molecule. Existing methods to compute, measure, and visualize the cavities in a protein molecule are sensitive to inaccuracies in the empirically determined atomic radii. We present a topological framework that enables robust computation and visualization of these structures. Given a fixed set of atoms, cavities are represented as subsets of the weighted Delaunay triangulation of atom centres. A novel notion of (ε, π)-stable cavities helps identify cavities that are stable even after perturbing the atom radii by a small value. This approach is used to identify potential pockets and channels in protein structures.
Method
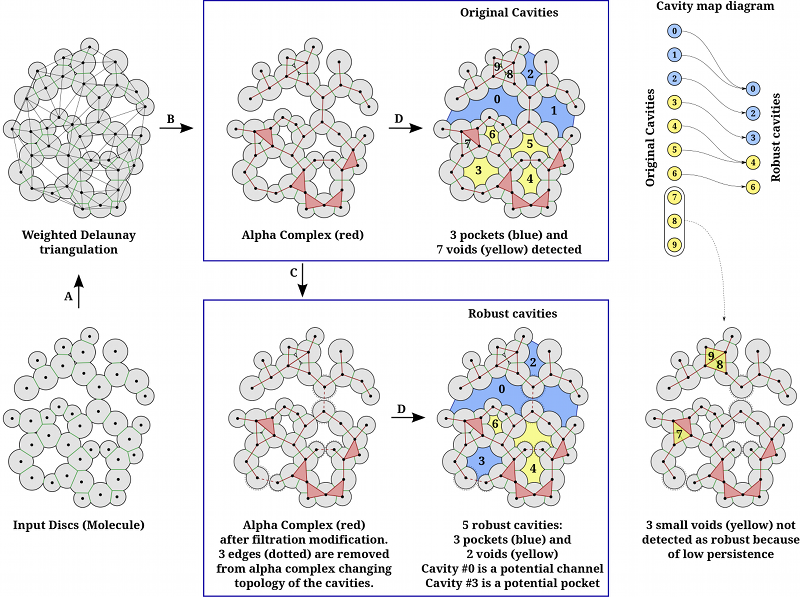
Graphical Abstract: The illustartion of extraction of robust cavities in a 2D example.
| Algorithm | |
| Input: | Molecule (as a set of spheres), ε, π. |
| Step A: | Compute weighted Delaunay triangulation of the set of spheres representing the molecule. |
| Step B: | Compute Alpha complex at α=0. |
| Step C: | Depending on value of ε, identify edges (triangles in 3D) in alpha complex which can be safely moved to the end of the filtration. The consequence of this step is that nearby cavities get merged into single cavity. |
| Step D: | Identify connected components in complement of the alpha complex to determine the Robust cavities. This step can also be applied before applying step C to obtain the Original Cavities. |
| Cavity pruning: | Robust cavities are pruned further based on topological persistence. This is controlled by parameter π, which removes cavities having persistence below π. This helps in removing insignificant cavities. |
| Definition of terms | |
| Cavity Map Diagram: | We compute cavities both before and after applying Step C. Thus, we have two sets of cavities called Original Cavities (before Step C) and Robust Cavities (After Step C and Cavity pruning). The mapping between these sets is represented as a cavity map diagram (shown on top right of the figure). |
| Cavity: | Maximally connected empty region within a molecule. |
| Void: | A buried cavity. It has no mouths (openings to molecular exterior). |
| Pocket: | A cavity with openings, i.e. number of mouths > 0. |
| Channel: | A cavity with at least two openings, i.e. number of mouths > 1. |
| Potential Channel: | A robust cavity with more than one mouths which is formed as a result of merging of cavities, each having at most one mouth. |
| Potential Pocket: | A robust pocket which has at least one void among its constituent. |
Use cases
Our method helps identify potential pockets and channels in protein which is not accomplished by traditional cavity computation algorithms. By potential pocket, we mean a void which can 'open up' by minor perturbation in atomic radii. Similarly, potential channel is defined as a set of nearby pockets which can merge after minor perturbation to form a channel structure.
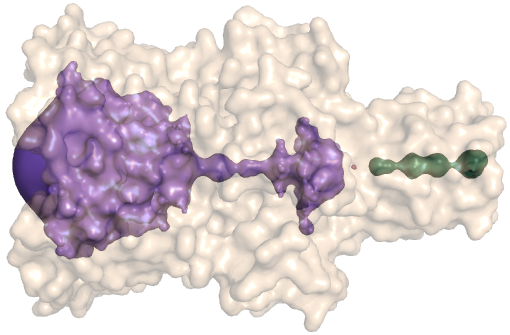
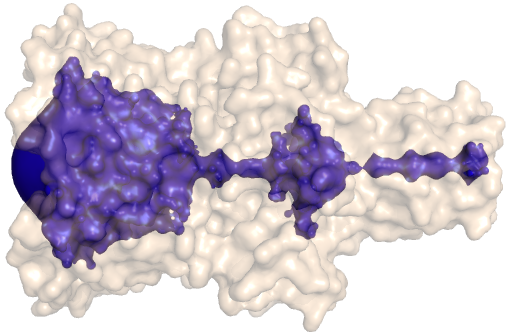
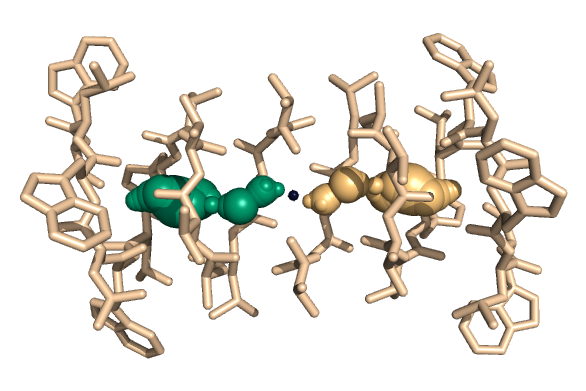
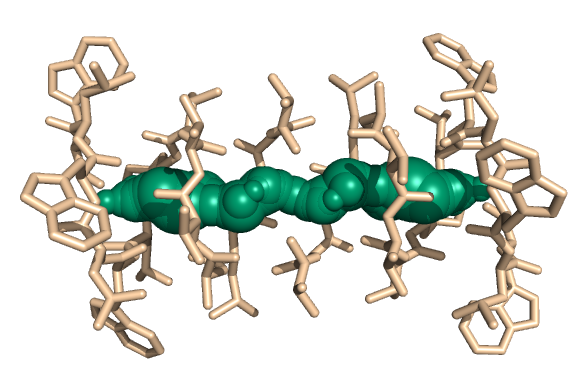
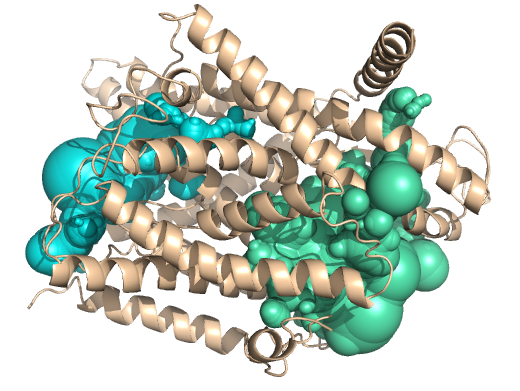
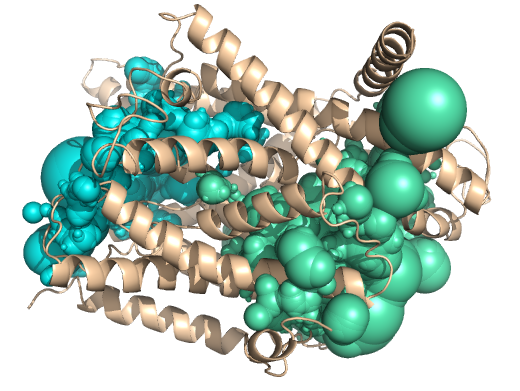
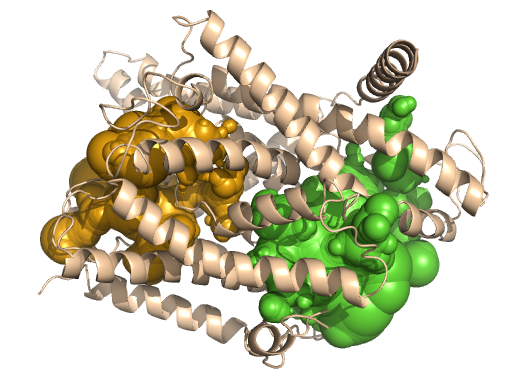
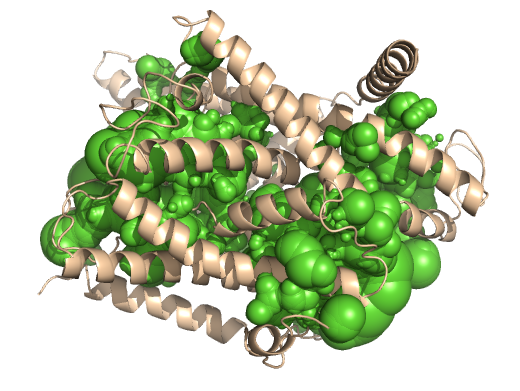
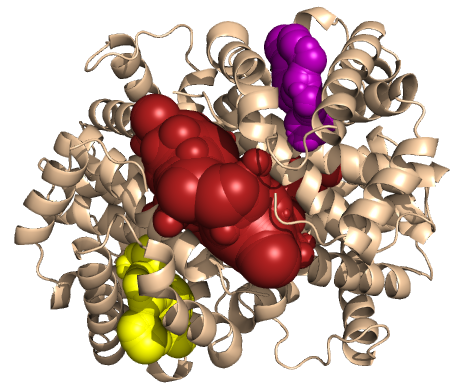
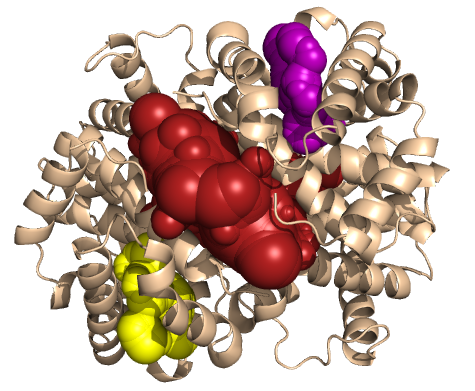
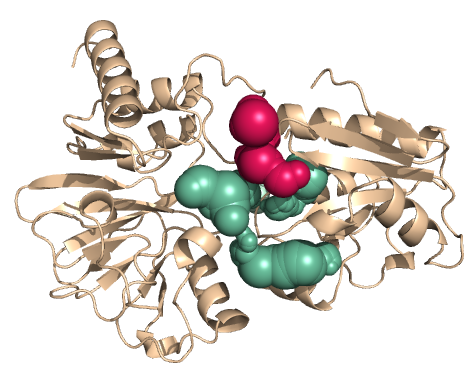
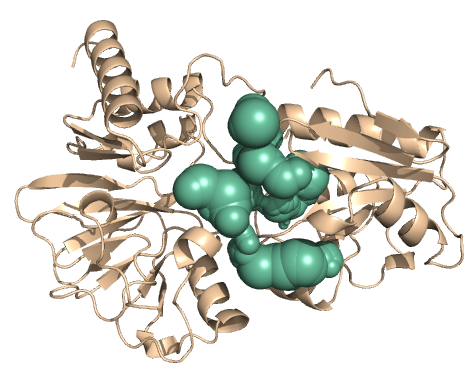
PDB-id: 2OAR — Left: Cavities detected by traditional algorithm, Right: RobustCavities successfully detects the potential channel.
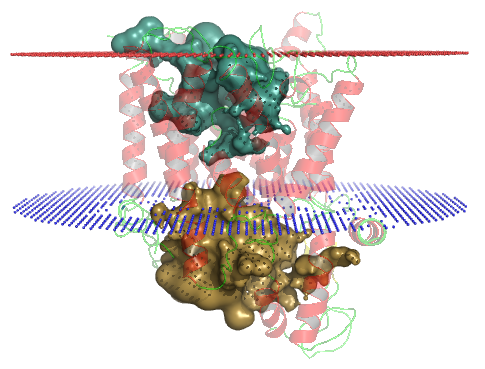

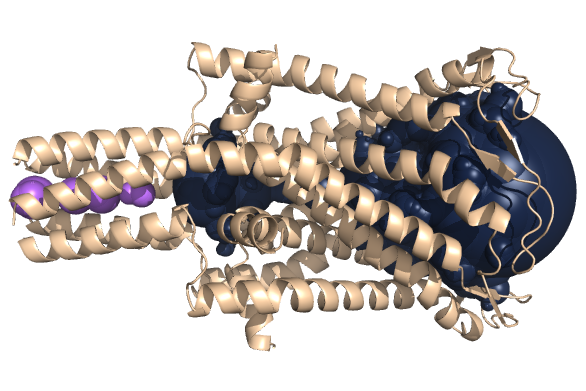
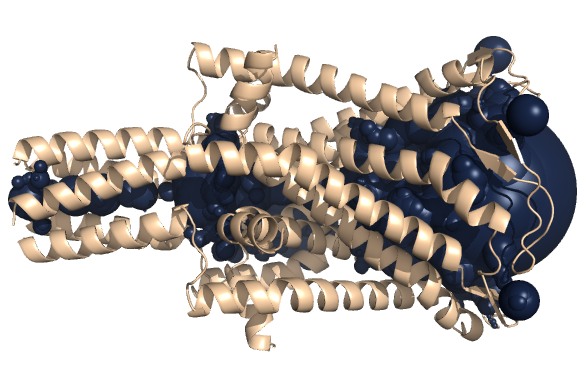
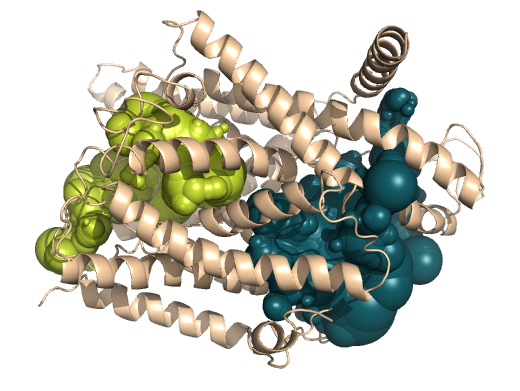
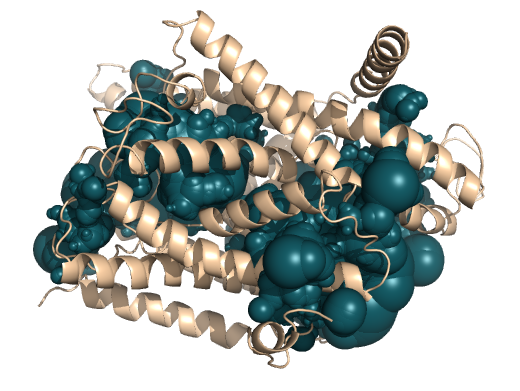
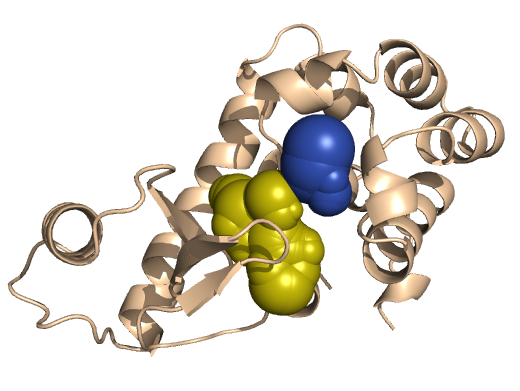
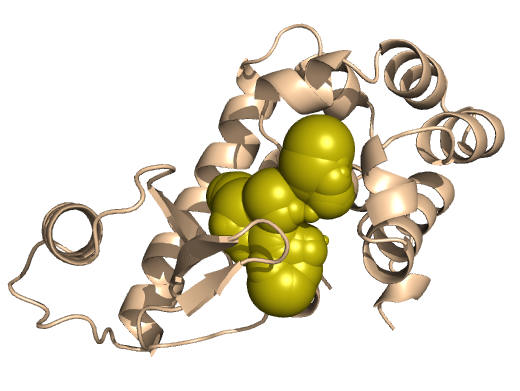
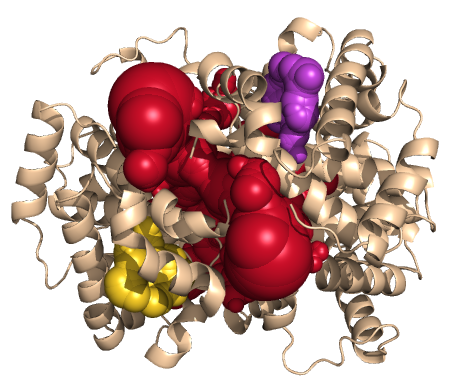
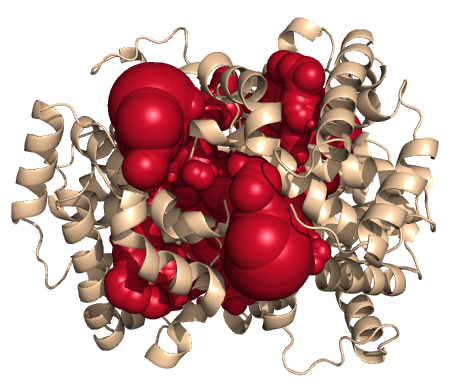
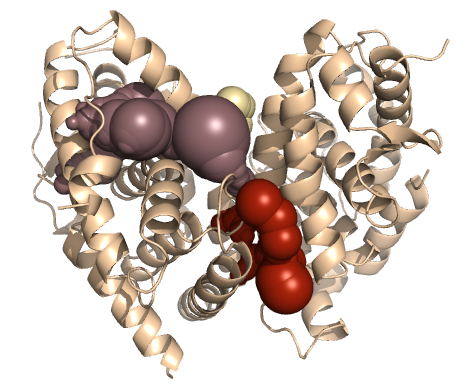
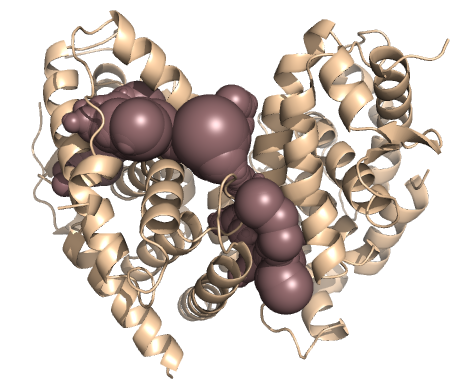
PDB-id: 2YXR — Left: Cavities detected by traditional algorithm, Right: RobustCavities successfully detects the potential channel.
References
- Raghavendra Sridharamurthy, Harish Doraiswamy, Siddharth Patel, Raghavan Varadarajan and Vijay Natarajan. Extraction of robust voids and pockets in proteins. EuroVis 2013: Eurographics Conference on Visualization (Short Paper), 2013. [pdf]
- Raghavendra Sridharamurthy, Harish Doraiswamy, Siddharth Patel, Raghavan Varadarajan and Vijay Natarajan. Extraction of robust voids and pockets in proteins. Technical Report, May 2013. [pdf]